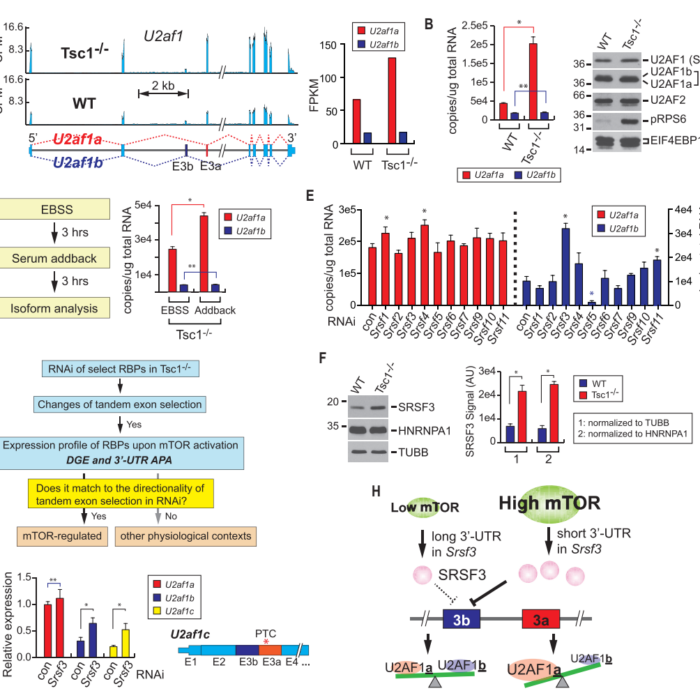
Our lab is interested in developing sophisticated machine learning approaches to extract useful information from the large-scale multi-omics data to understand the complex disease such as cancer. Our research covers several important topics in cancer transcriptome, spanning from technique-driven research that aims at developing graph-based learning models for cancer transcriptome analysis with prior knowledge (e.g., isoform quantification, biomarker identification, cancer outcome prediction, drug sensitivity prediction), to hypothesis-driven investigation of specific biological problems (e.g., changes of transcriptome upon mTOR hyper-activation). Our development leads to novel computational models and molecular signatures, which could be used in early detection, diagnosis, and prognosis of specific tumors.
List of Projects
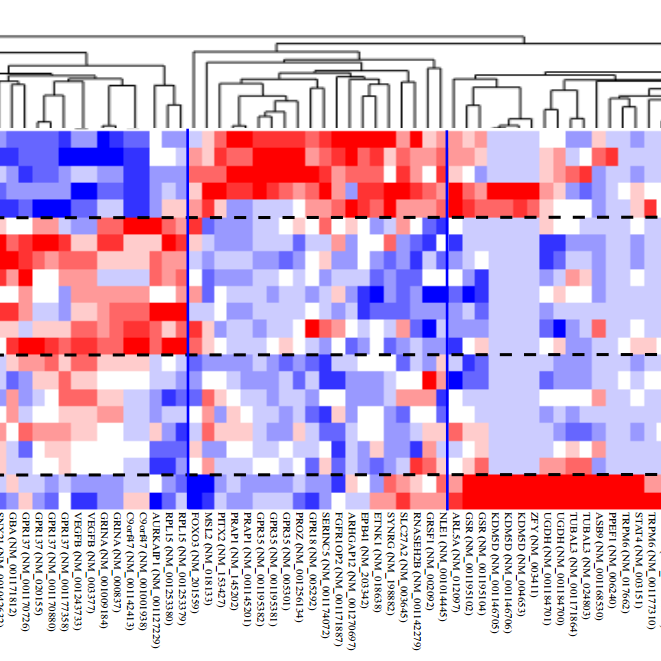
omicsGAT
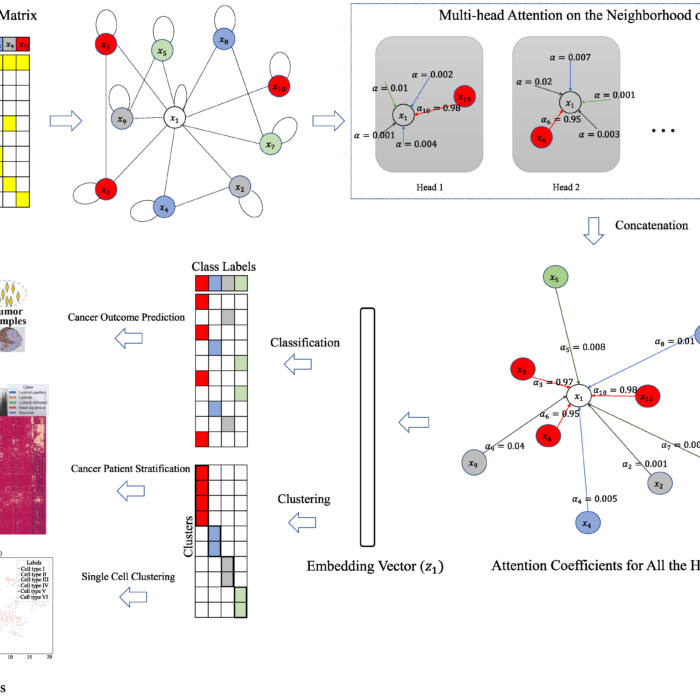
Graph Attention Network for Cancer Subtype Analyses
CancerCellTracker
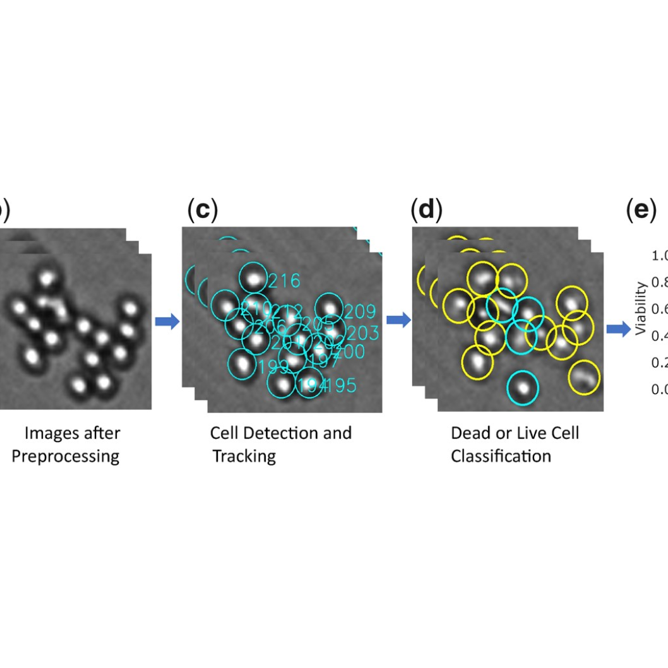
A Brightfield Time-lapse Microscopy Framework for Cancer Drug Sensitivity Estimation
APA-Scan
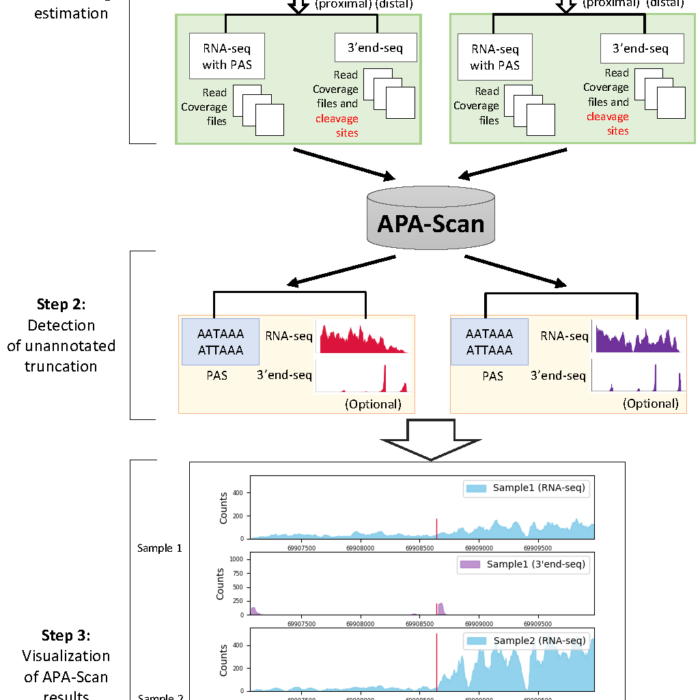
Detection and Visualization of 3′-UTR APA with RNA-seq and 3′-end-seq Data
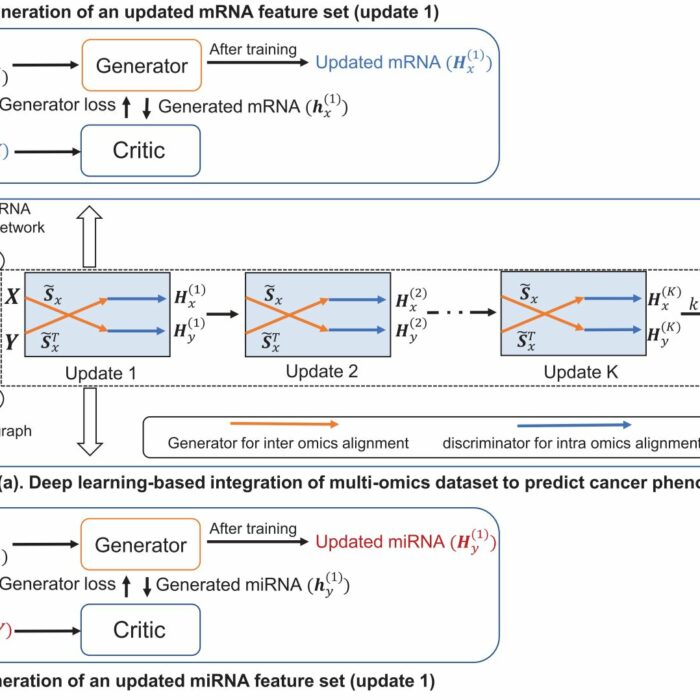
omicsGAN
Multi-omics Data Integration by Generative Adversarial Network
PTNet
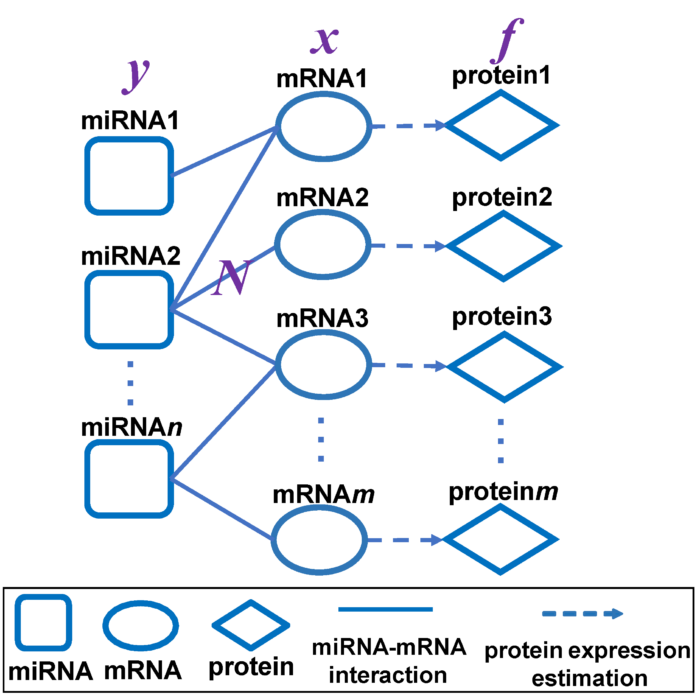
In silico model for miRNA-mediated regulatory network in cancer
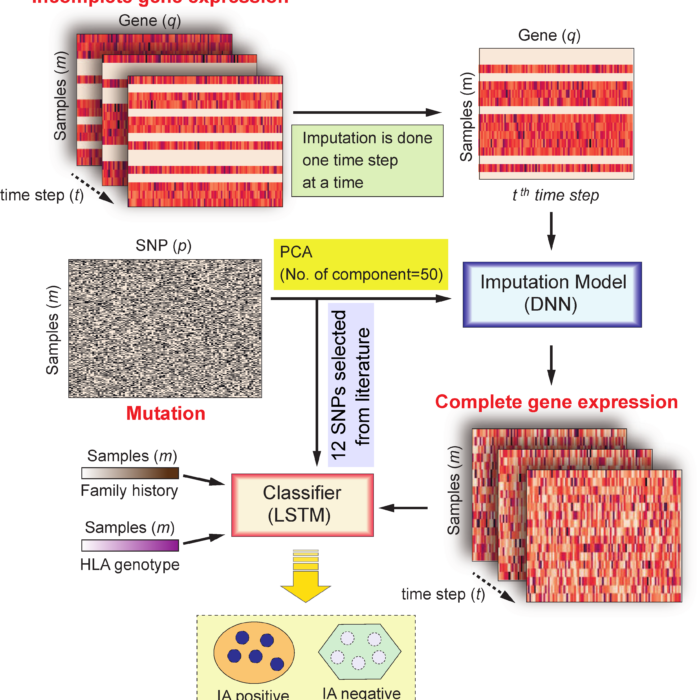
Incomplete Time-Series Gene Expression in Integrative Study
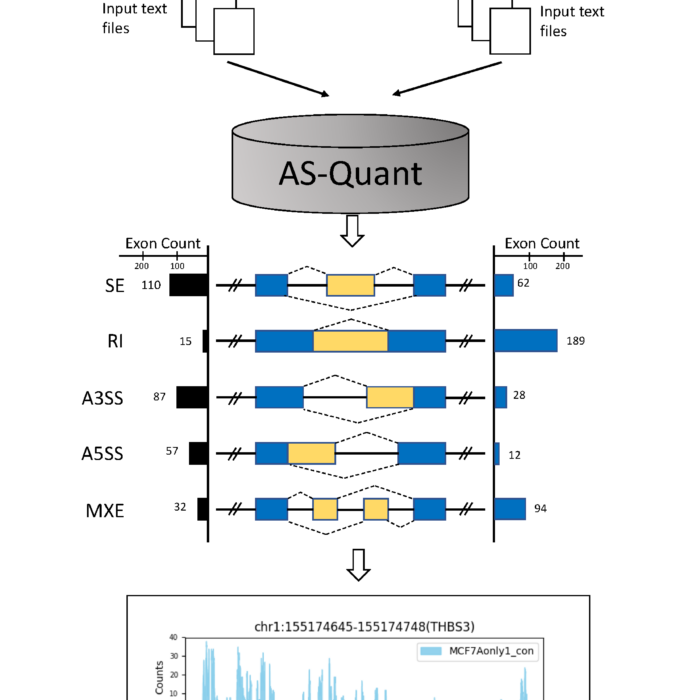
AS-Quant
Detection and Visualization of Alternative Splicing Events with RNA-seq Data
Graph-based Drug Sensitivity Prediction
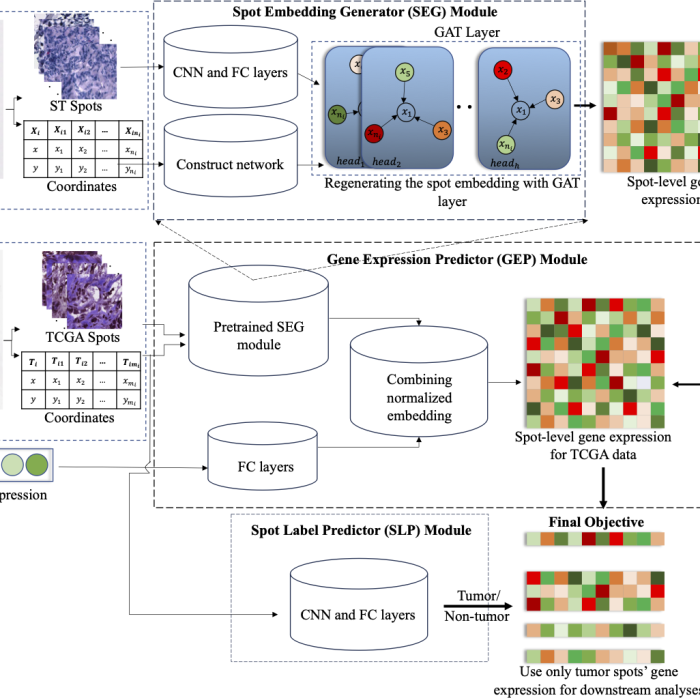
PTNet
Platform-integrated mRNA Isoform Quantification
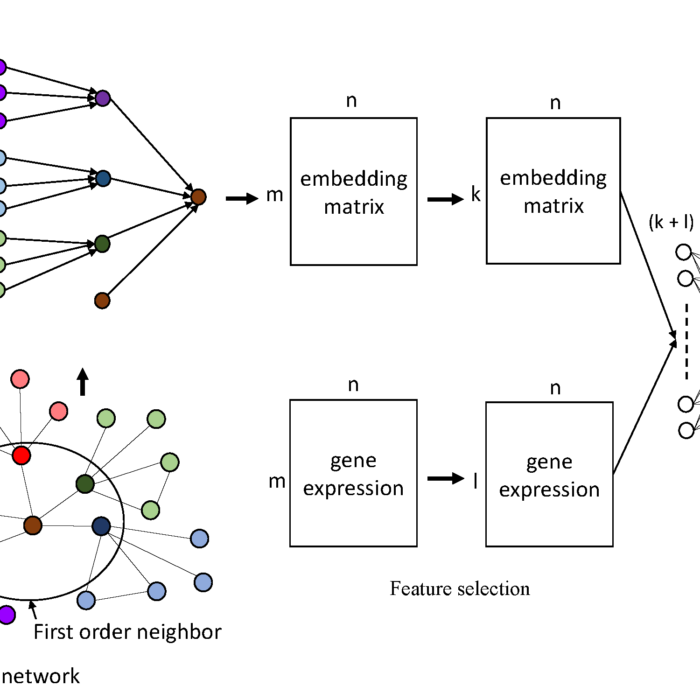
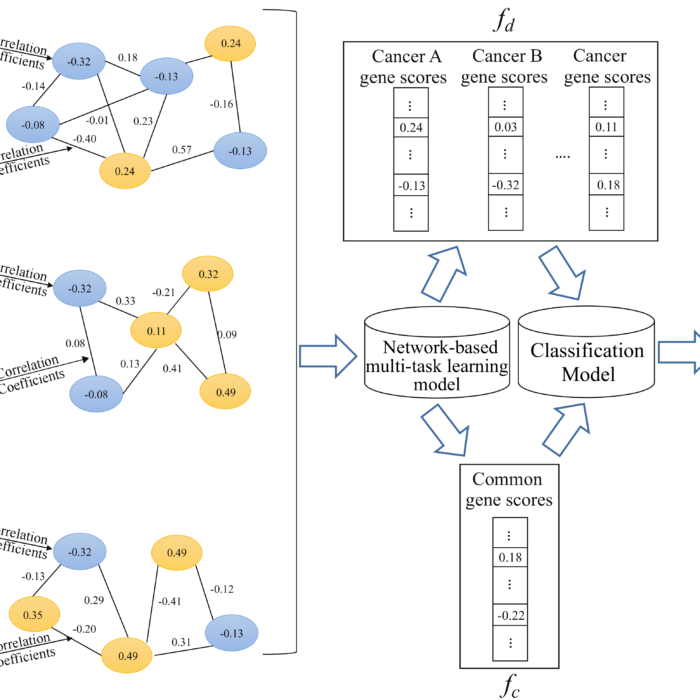
NetML
Network-based Multi-Task Learning Models for Biomarker Selection and Cancer Outcome Prediction
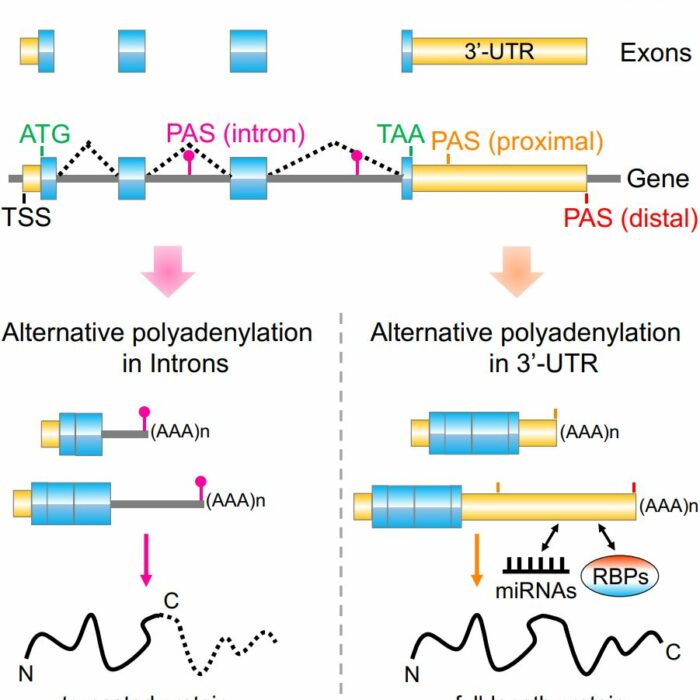
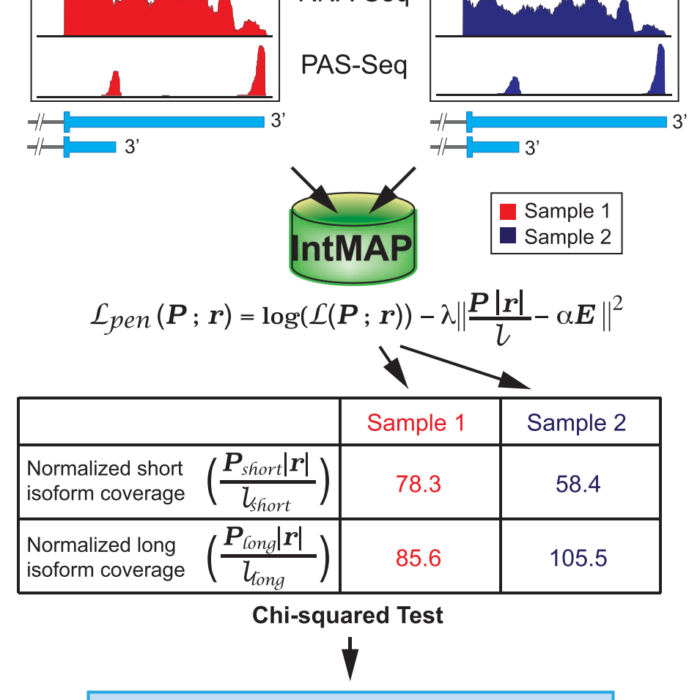
IntMAP
An Integrative Model for Alternative Polyadenylation